Stallaert Lab
Clarifying complex systems
What are we interested in?
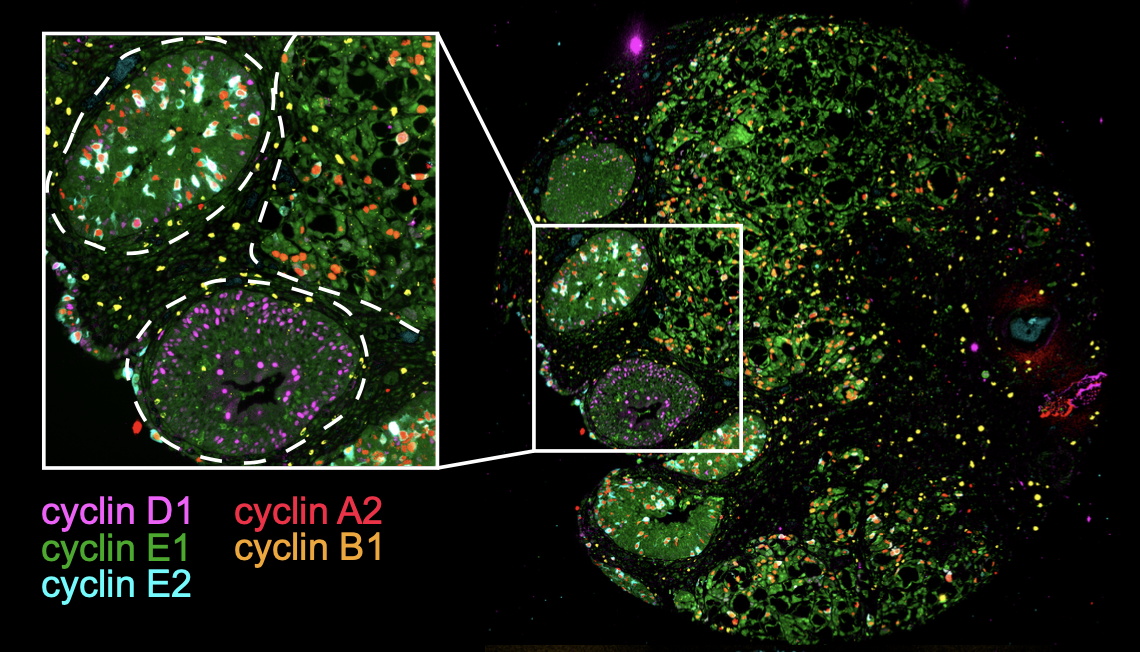
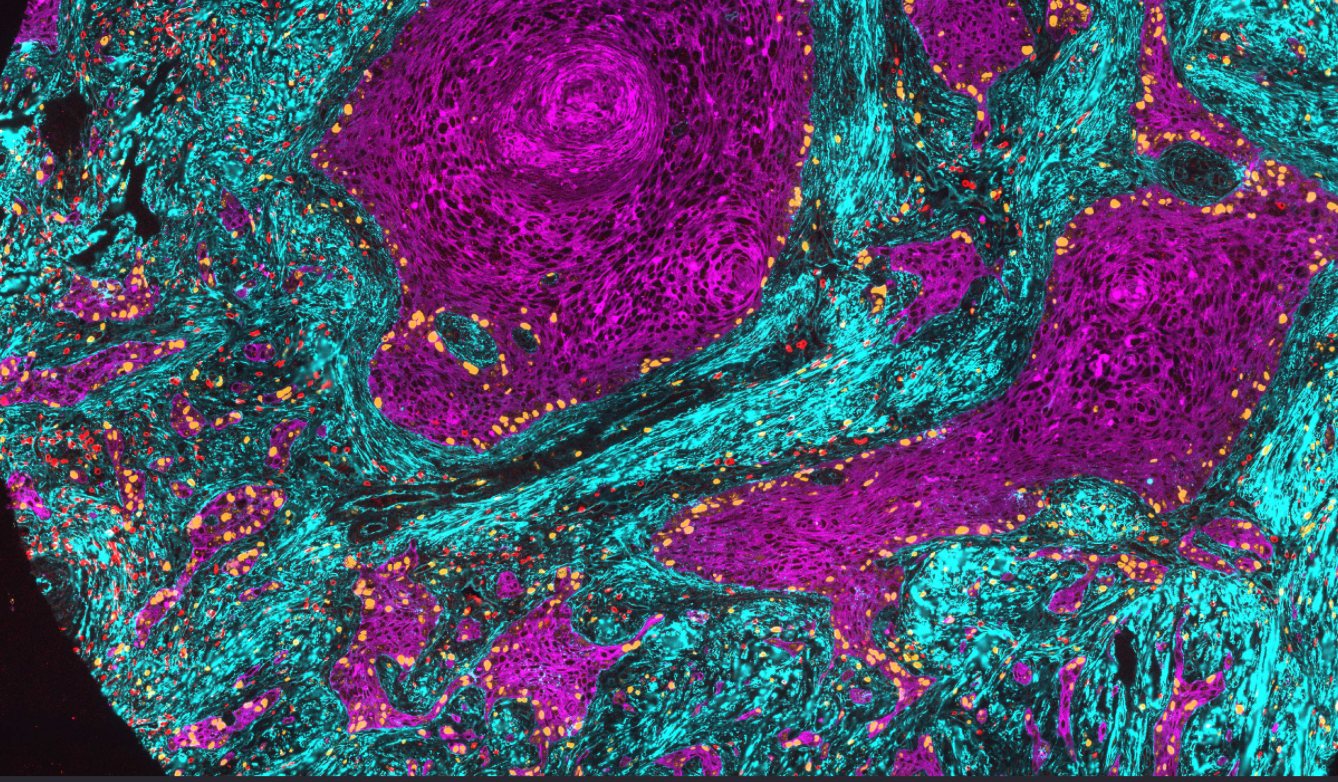
Studying the cell cycle in its native environment
Investigating the role of the TME in cancer cell proliferation
The tumor microenvironment (TME) plays a major role in the initiation, progression, and drug sensitivity of tumors. However, we know very little about how the TME influences cancer cell proliferation and arrest. Does the TME expand or limit the repertoire of cell cycle paths available to a tumor cell? Using tissue specimens of patient tumors, we are trying to understand how the TME influences the proliferation/arrest decision of cancer cells.
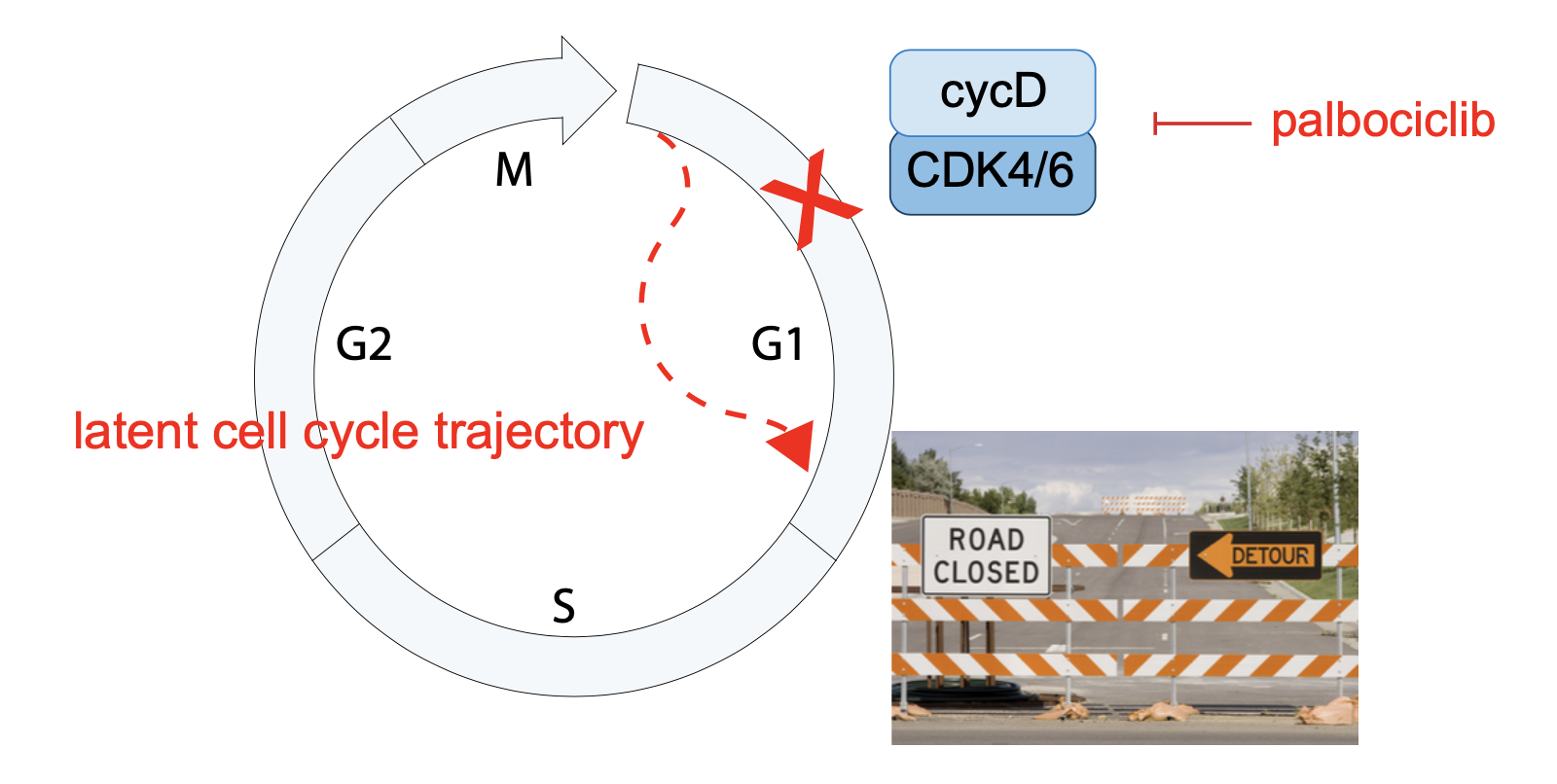
Developing precision medicine that target tumor-specific cell cycles
With the advent of technologies that allow us to measure cell cycle progression at the single-cell level, we now know that cells do not always traverse the same mechanistic paths en route to cell division. To explore this plasticity in cell cycle regulation, we are mapping the cell cycles of various cell types embedded in their native, 3-dimensional tissue environment to better understand the extent and functional role of diverse proliferative programs in human physiology and disease.
Cancer is a disease characterized by uncontrolled cell proliferation and many of the mutations that drive tumorigenesis occur directly in the proteins that control the cell cycle. Using tumor microarray libraries, patient-derived organoids, and cell line models, we study how the cell cycle adapts during tumor initiation, progression, and with drug treatment, with the ultimate goal of identifying specific vulnerabilities in the cancer cell cycle and targeting them with personalized therapy.
How do we do it?
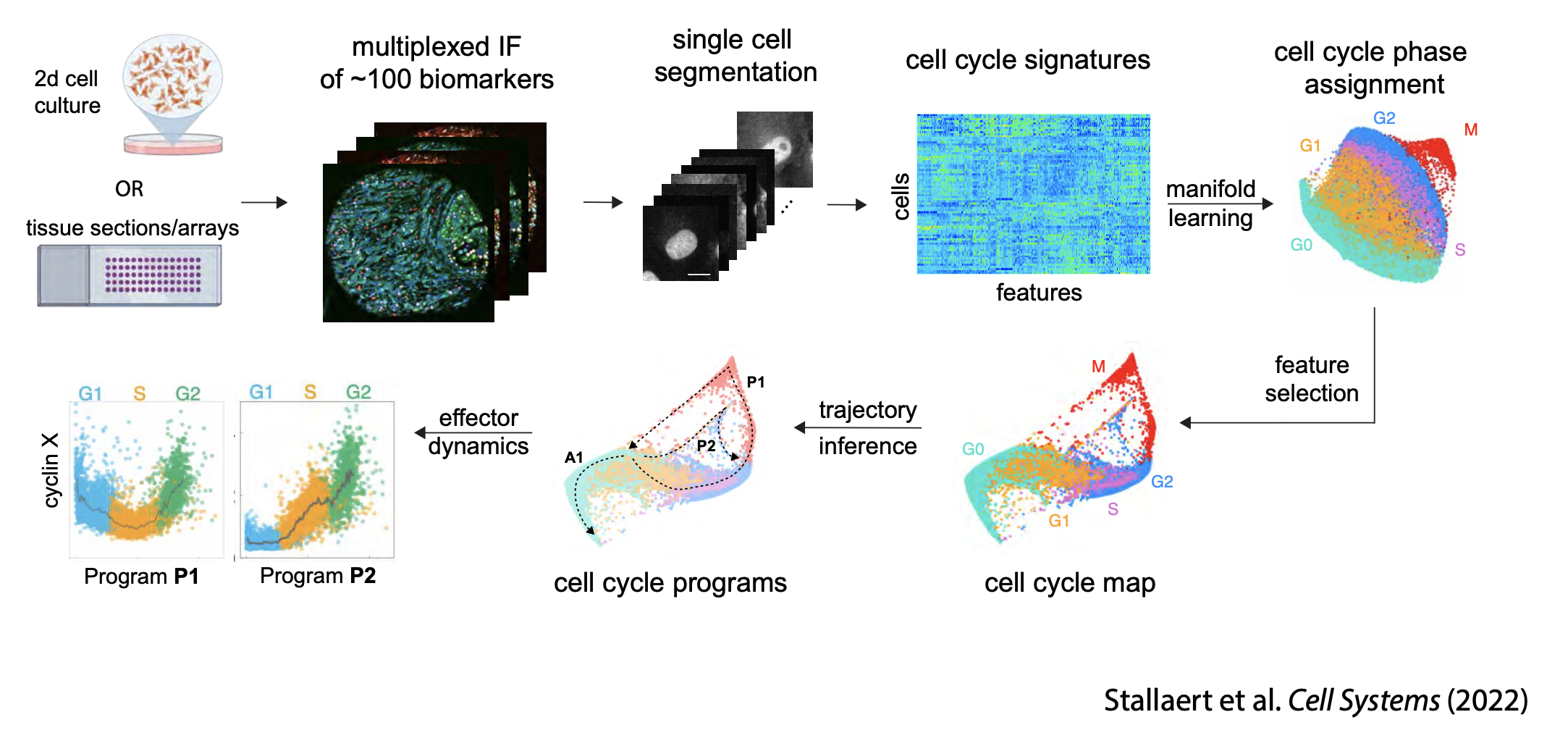
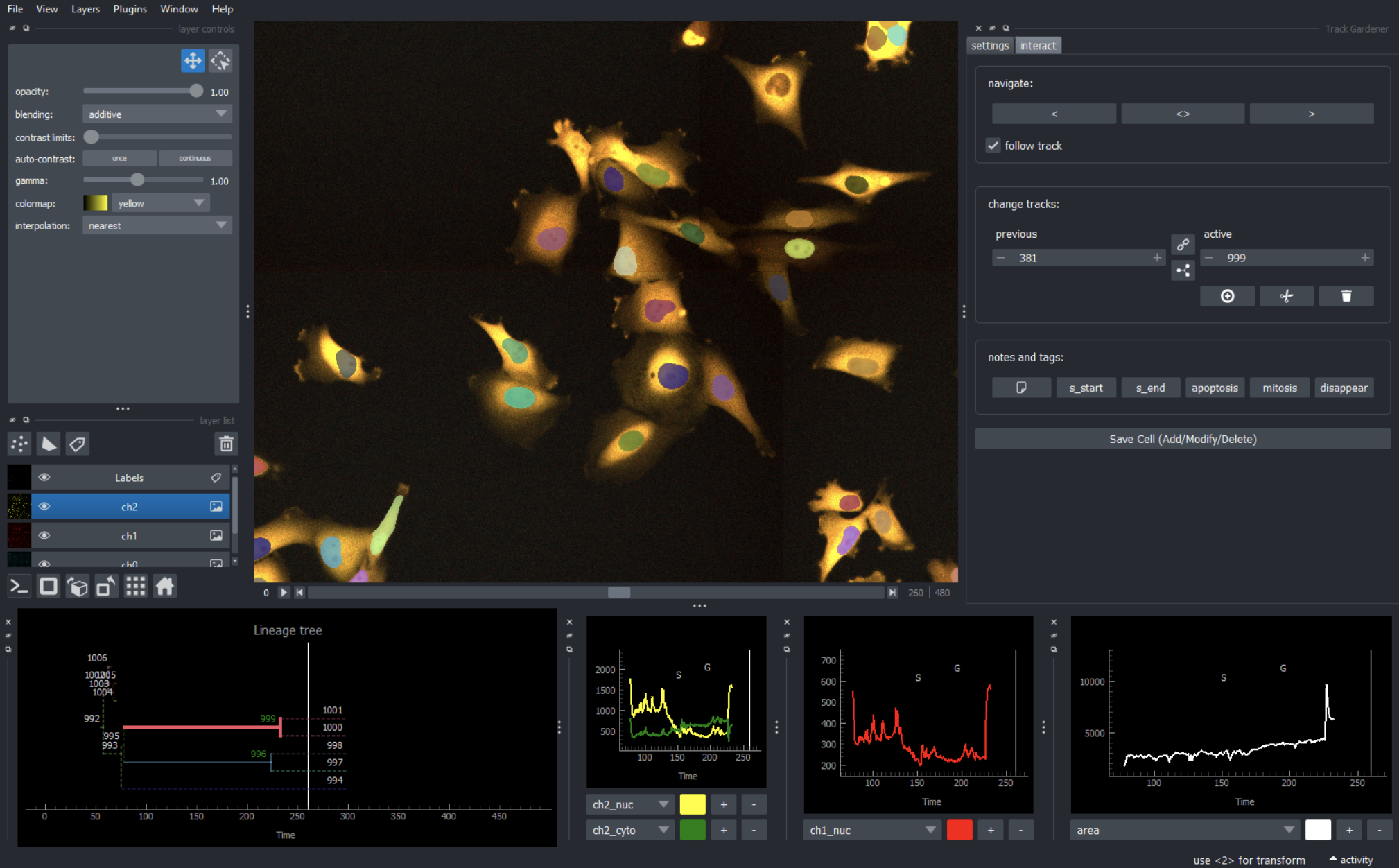
Cell cycle mapping
We combine hyperplexed, single-cell imaging with manifold learning to map the diversity of paths that cells take through the cell cycle
Live-cell imaging
Using fluorescent biosensors of signaling and cell cycle effector activity we track individual cells as they progress through the cell cycle